Mapping Regional Astrocyte Transcriptional Dependencies in Brain Circuits
Our studies on astrocyte diversity revealed that several of the transcription factors (TFs) that regulate glial development during embryogenesis are also expressed in mature astrocytes in the adult brain. Among these TFs is Nuclear Factor I-A (NFIA) and Sox9, genes that my lab has studied in the embryonic spinal cord for many years, however whether and how they contribute to mature astrocyte function in the adult brain is unknown which led us to examine their roles in mature astrocytes in the adult brain.
Despite exhibiting universal expression in nearly every astrocyte and across all brain regions, we discovered that NFIA is selectively required for hippocampal astrocyte function. Loss of NFIA resulted in reduced morphological complexity and Ca2+ activity in the hippocampus alone and not the cortex, olfactory bulb, or brainstem. These alterations in astrocyte function are combined with diminution of learning and memory behaviors and inhibition of long-term potentiation (LTP). These studies discovered astrocyte region-specific transcriptional dependencies and identified astrocytic NFIA as a key transcriptional regulator of hippocampal circuits (Huang, et al. Neuron 2020). Using parallel approaches for Sox9, we found that it is specifically required to maintain the morphology of olfactory bulb astrocytes and olfactory circuit function through regulation of GLT1 expression (Ung, et al. Nature Communications 2021). Finally, our recent developmental studies revealed that these transcriptional dependencies for NFIA and Sox9 in the adult are developmentally encoded and intertwined with activity dependent processes via GABA signaling (Cheng, et al. Nature 2023). These studies highlight the important role of TFs in astrocyte function, which we are active areas of investigation.
Experience-Dependent Astrocyte Plasticity
The extent to which astrocyte function is influenced by experience and how these alterations contribute to circuit adaption and plasticity after sensory or environmental experiences remains very poorly defined. To understand how astrocytes adapt to environmental experiences, we used a juvenile social deprivation paradigm in mice.. We found that astrocyte function is tuned to social experience and identifies a social context specific mechanism by which astrocytic TRPA1 and GABA coordinately suppress hippocampal circuit activity and cognitive function (Cheng, et al. Neuron 2023). In a parallel study, using olfactory sensory experience, we found that neuronal activity and odor exposure induces widespread changes in gene expression in astrocytes, highlighted by drastic changes in Sox9-DNA binding. Mechanistically, we discovered that serotonin transporters are induced in astrocytes after odor exposure and regulate histone serotonylation, a new form of epigenomic regulation. Perturbation of histone serotonlyation in astrocytes suppresses olfactory responses and associated circuit activity (Sardar, et al. in final review).
In broad terms, these studies describe how information exchanged from neurons to astrocytes is integrated, encoded, and utilized to shape circuit activities. The intersection of neuronal activity and epigenomic remodeling in astrocytes in intact circuits represents a new form of astrocyte-neuron communication and further highlights the complexity of astrocyte plasticity in response to experience. The tools, systems, and concepts described from these studies will serve as a foundation for additional investigations into astrocyte adaptation to experience.
Cancer Neuroscience: Signaling Between Brain Tumors and Neurons
A series of recent studies have shown that neuron to glioma signaling plays an important role in tumor progression and pathophysiology. Studies from our lab complement- and expand upon these findings by showing that glioma tumors promote neuronal activity, thus propagating a vicious cycle of tumor to neuron crosstalk that culminates in glioma progression and brain network hyperactivity resulting in glioma induced-epilepsy. Moreover, these studies illustrate that glioma cells release factors that remodel neuronal synapses towards hyperactivity (Yu, et al. Nature 2020). Thus brain tumor highjack astrocytic functions to remodel neuronal synapses toward hyperactivity, driving glioma-related epilepsy.
Our more recent studies used intraoperative recordings from patients with glioma-related epilepsy and functional screening in mouse glioma models to identify new pathways and genes that promote glioma progression by interfering with K+ buffering, which facilitates brain network hyperactivity (Curry, et al. Neuron 2023). Finally, we expanded our research to examine neuronal regulation of glioma invasion, finding that neuronal projections at locations remote to the primary tumor specifically drive glioma infiltration through the upregulation of axon guidance genes in glioma tumors (Huang-Hobbs, et al. Nature 2023).
Functional Genomics of Malignant Glioma
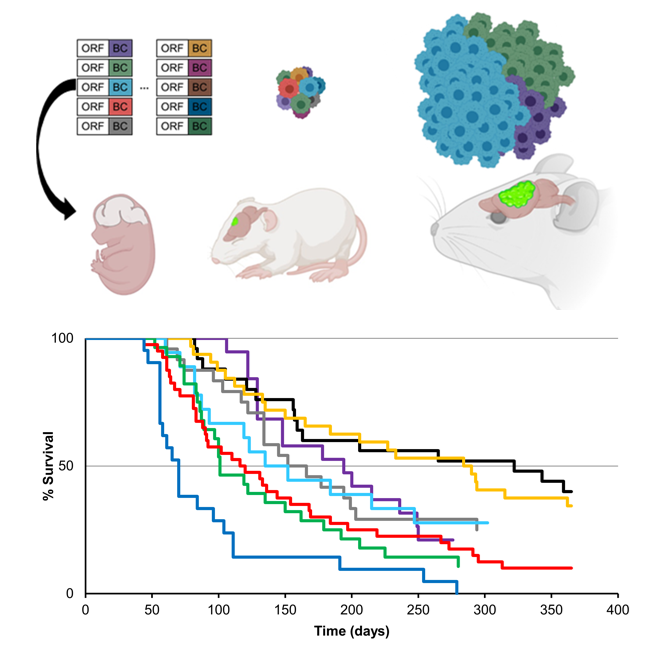
Over the past 20+ years, cancer genomics has cataloged nearly every tumor-associated mutation. Although such crucial advances hold the promise of personalized cancer treatments, one major obstacle in achieving this goal is decoding driver and passenger mutations from cancer cohorts. To date, amongst the over 3 million cancer mutations identified, only about 5,000 have been functionally annotated. In collaboration with Gordon Mills and our CTD2 team, we employed our CRISPR-based, autochthonous mouse model of glioma to develop an in vivo functional genomics screening platform to identify novel driver variants of key oncogenes in glioma. Screening an allelic series of PIK3CA variants present in human glioma, we identified a novel PIK3CA driver variant of glioma tumorigenesis (Yu, et a. Nature 2020). Moreover, we performed a parallel allelic screen with EGFR variants and identified a series of novel EGFR driver variants in glioma (Yu, et al. NeuroOncology 2023).
In collaboration with Melissa Bondy, we expanded our functional genomics screens to Familial Glioma, screening a series of gene variants that are associated with an increased risk of developing glioma (Choi, et al. Science Advances 2023). Finally, we utilized our in vivo, functional genomics screening platform in our Cancer Neuroscience studies to identify key regulators of brain tumor-induced epilepsy and infiltration (Huang-Hobbs, et al. Nature 2023; Curry, et al. Neuron 2023)